University of Pennsylvania |
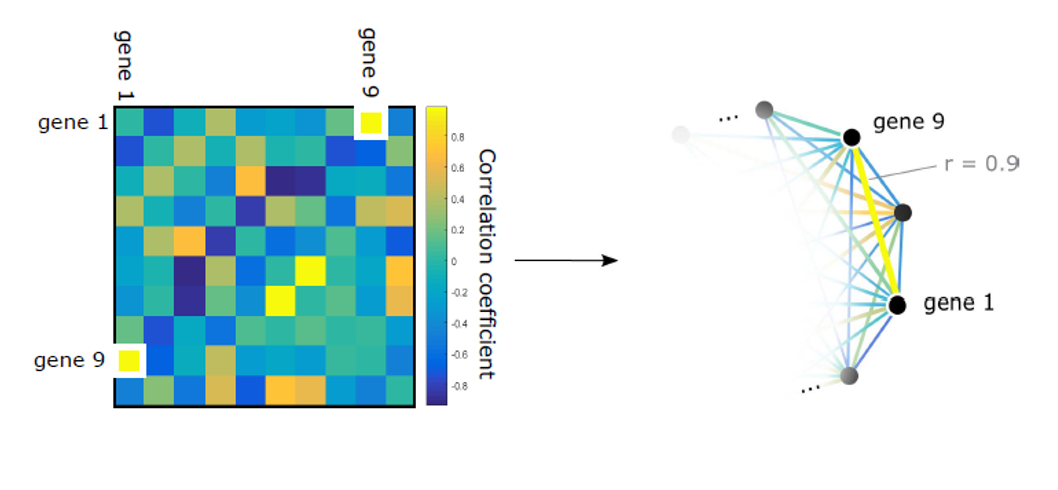
Encoding neural wiring and activity as a network welcomes the application of various tools from theoretical mathematics in the analysis of brain data, thus progressing the understanding of the structure-function relationship as well as the learning process. My work in the Complex Systems group focused on evolving mathematical techniques for computational network analysis. I then used these methods to determine a classification of previously reported models based on their structural features and considered the effect of such architecture on network function. The code and reference for this work can be found on the Network Toolbox page. In December 2015 I completed a Master's thesis on this work, titled "Cliques and Cycles: A Complementary Pair of Homological Features Detects Structure in Weighted Networks."
|
Broad Institute of Harvard and MIT Using a parallel loss-of-function screen, the Project Achilles team evaluates the effect of gene loss in multiple cancer cell lines. As an associate computational biologist I worked on the data processing and analysis for this program. My research projects included applying tools from network science to answer questions in cancer biology.
|
Stowers Institute for Medical Research |
Transport of proteins across the nuclear membrane is essential for cell function and viability. Alternate sets of secretory proteins may form complexes depending on their position in the contiguous membrane and presented task. As a summer scholar in the Jaspersen lab, I created strains of Saccharomyces cerevisiae containing tagged proteins and used confocal microscopy techniques to study their interactions along the nuclear membrane and endoplasmic reticulum. I presented findings at Summer Scholars Poster Session in August 2014.
|
Boston College Aspartate transcarbamoylase (ATCase) from Escherichia coli is the main regulator in the de novo pyrimidine nucleotide biosynthesis pathway. ATCase follows the Monod, Wyman, Changeux Model for Allostery, regulated by nucleotide triphosphates. As an undergraduate I worked with a regulatory mutant of ATCase beginning first with the growth and purification of the enzyme followed by creating a kinetic profile from a series of colorimetric assays. Additionally I integrated these results with Small Angle X-ray Scattering results and three crystal structures of the mutant from X-ray diffraction data solved using Phenix and COOT. In summary, this work further delineated the path of information flow from the allosteric to active site of wild-type ATCase.
|
Ann sizemore blevins
ann.e.sizemore@gmail.com